DECODR™
Biocomputing made easy and powerful
Accurate. Intuitive. Visual. CRISPR analysis at its full potential.
.jpg/_jcr_content/renditions/original)
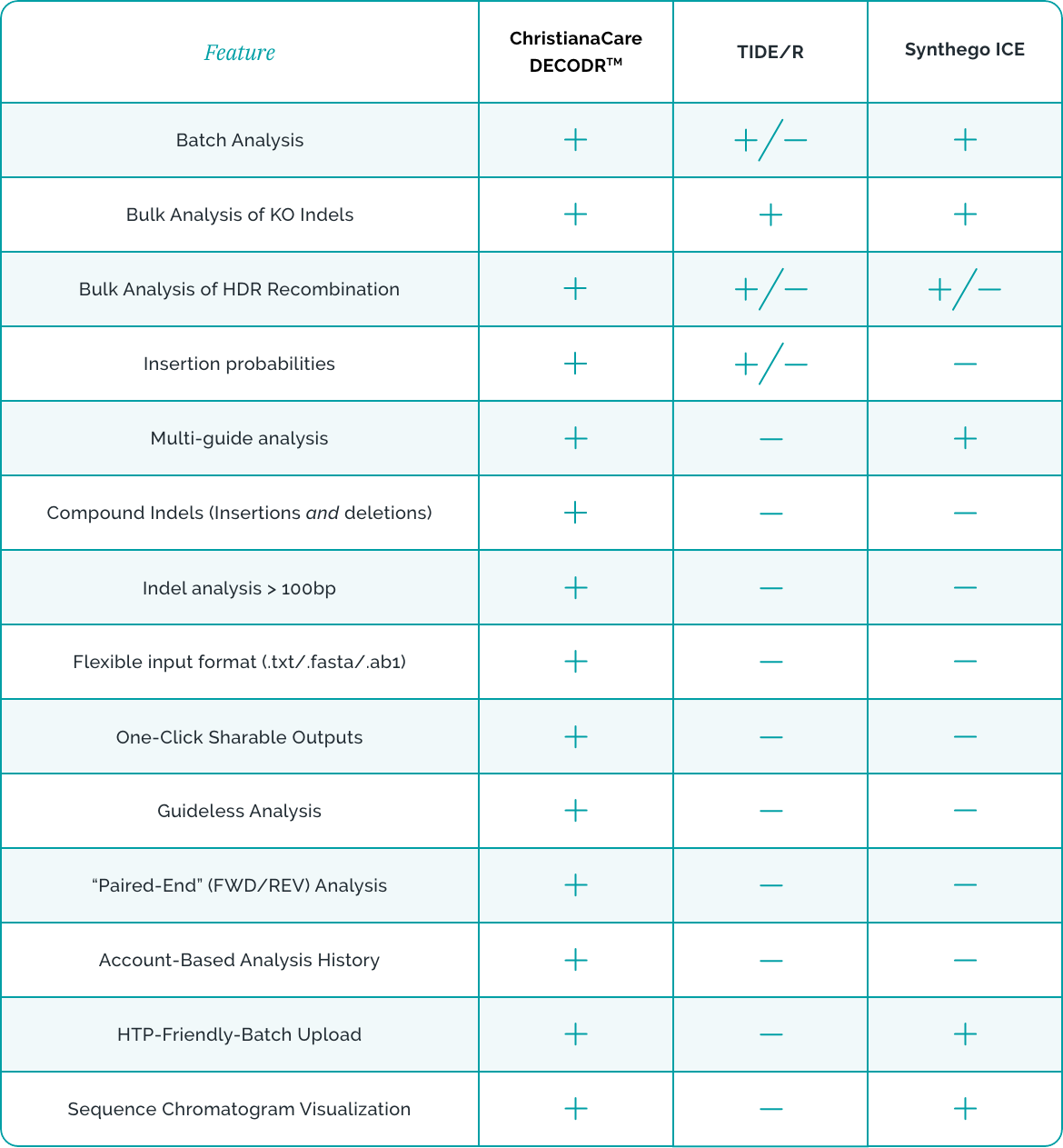
DECODR™ (Deconvolution of Complex DNA Repair) provides a convenient and user-friendly method to quantify the edits present in CRISPR-edited Sanger sequencing data. DECODR™ has been recognized in Nature Scientific Reports as the best tool to evaluate CRISPR edits for somatic in-vivo gene editing, as it does not have a limit on the length of indels it measures.
DECODR™ uses a computationally efficient algorithm and provides the identity of deleted and inserted bases with no limit on insertion and deletion size. DECODRTM supports batch analysis, in which several sequencing files can be analyzed simultaneously.
To use DECODRTM, all you need are Sanger sequencing files from your experimental samples. You upload the sequence files to DECODRTM via drag-and-drop and provide experimental details. DECODR will provide you with NGS-level data output about your CRISPR edits in just seconds.
"DECODR™ is an indel analysis tool that has comparable deconvolution methods as TIDE/Synthego, but the lack of a window limit makes DECODR™ better equipped for somatic in-vivo gene editing." - Nature Scientific Reports
For more information about DECODRTM , see our publication in The CRISPR Journal and Nature Scientific Reports.
DECODRTM is available to all non-profit and academic researchers. For-profit users and commercial interest in DECODR can contact us at licensing.decodr@geneeditinginstitute.com
Please visit DECODR.org to register your account and start using DECODR in your lab today.
Need more information?
To find out more about DECODR, email us at decodr@geneeditinginstitute.com (Non-profit & Academic Interest) or licensing.decodr@geneeditinginstitute.com (For-profit & Commercial Interest).